OVO Database¶
Data model¶
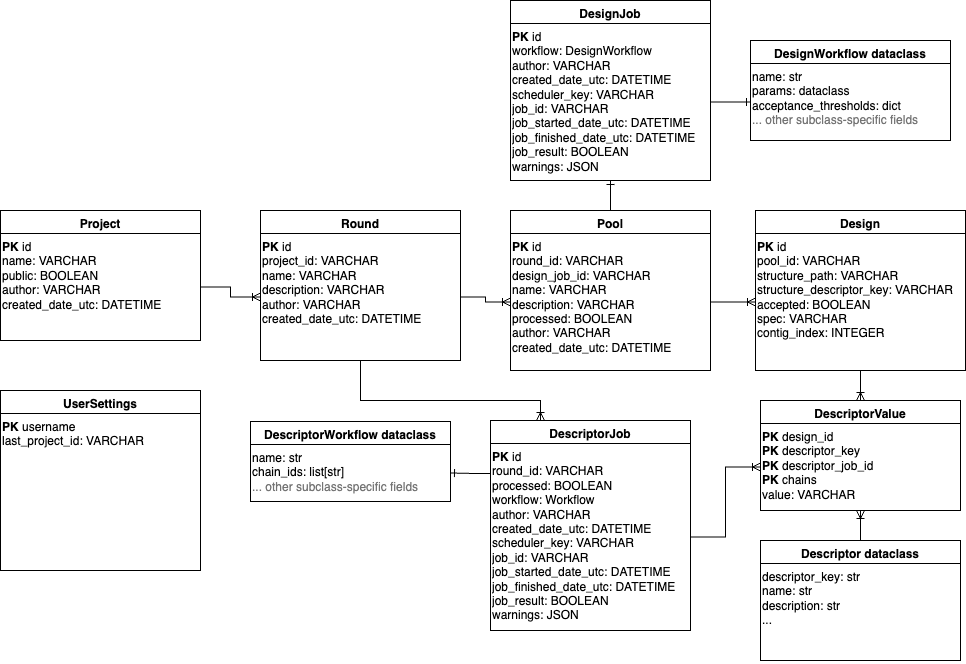
In the OVO data model, a Design corresponds to one designed molecule defined by its sequence.
Designs are organized into Pools, where each Pool is created by a single submitted workflow
or uploaded by the user.
Submission parameters and job status metadata are stored in a separate DesignJob model
using a workflow field storing a JSON-serialized instance of a DesignWorkflow dataclass.
The DesignWorkflow dataclasses define workflow parameters and encapsulate use-case-specific logic.
New use-cases can be added by OVO plugins that implement their own DesignWorkflow subclasses.
Each Pool is identified by a randomly generated three-letter code (such as qki) that is
unique within the current database instance. This ID is used as a prefix for the ID of each Design
along with the backbone and sequence identifier (such as ovo_qki_0045_cycle01), ensuring that each
design can be uniquely tracked to its job of origin (and the corresponding workflow parameters).
Pools can be grouped into Rounds, corresponding to the iterative design-make-test-analyze loop,
and further into Projects, enabling sustainable organization in a multi-user setting.
Each design can be annotated with descriptors using the DescriptorValue model, which
is a key-value “tall” representation where all descriptor values are stored in a single
value column and associated with the descriptor using a descriptor_key column
(as opposed to a “wide” representation which would use a separate column for each descriptor).
Each DescriptorValue row is a single value associated with a single descriptor_key,
Design and DescriptorJob, storing workflow parameters in a DescriptorWorkflow dataclass, analogous to design jobs.
Different types of Descriptors (such as AlphaFold2 pLDDT, Rosetta ddG, or total positive patch area)
are uniquely defined by their descriptor_key and provide additional metadata such as their
human-readable name and description. The Descriptor type objects are not defined in the database
but inside OVO codebase and extended by plugins. Protein structures and other files are not stored
directly in the database but referenced by their storage path as managed by the Storage class.
The structure path string can be stored as a field of the Design model or as a special type of Descriptor.
Interacting with the database¶
Database rows can be retrieved from the database using the db object using select and get methods:
from ovo import db
# List all projects
db.Project.select()
# [Project(author='username', created_date_utc=datetime.datetime(2025, 11, 13, 9, 58, 36, 339174), id='b8e657bb-b0b0-423e-9235-383a6d8f74e5', name='OVO Publication Examples 1', public=True)]
# Get project by name
project = db.Project.get(name="OVO Publication Examples 1")
print(project.id)
# b8e657bb-b0b0-423e-9235-383a6d8f74e5
# Get Pool object
pool = db.Pool.get(id="qki")
print(pool.id)
print(pool.name)
# qki
# My first design pool
# Get list of Design objects in the pool
all_designs = db.Design.select(pool_id="qki")
print(all_designs[0].id)
# 'ovo_qki_0001_cycle01'
# Get only accepted designs
accepted_designs = db.Design.select(pool_id="qki", accepted=True)
print(accepted_designs[0].id)
# 'ovo_qki_0125_cycle02'
Use the project_logic module to retrieve projects, rounds and pools:
from ovo import db, project_logic
project, round = project_logic.get_or_create_project_round(
"OVO Publication Examples 1",
"Round 1",
)
print(round.id)
Use the design_logic module for convenience functions to retrieve designs and pools:
from ovo import db, design_logic
project = db.Project.get(name="OVO Publication Examples 1")
pools = design_logic.get_pools_table(project_id=project.id)
pools.head()
# Round ID Name ...
# 0 Binder design bbc 4ZXB 1000 designs default weights ligandmpnn ...
# 1 Binder design qki Top designs diversification ...
# 2 Motif scaffolding jov 1A41 6*100*8 designs default weights ...
# 3 Interface scaffolding zuk 5IUS 500*5 designs PD1 interface ...
# 4 Binder design mmo 4ZXB 1000 designs beta sheet ...
Use the descriptor_logic module for convenience functions to retrieve descriptor values:
from ovo import db, descriptor_logic
# Get designs as a pandas DataFrame
designs = db.Design.select_dataframe(pool_id="qki", accepted=True)
designs.head()
# pool_id structure_path ...
# id ...
# ovo_qki_0045_cycle01 qki project/fd3f1fcc-1dda-4076-ad4e-7b885ec90032/p... ...
# ovo_qki_0045_cycle02 qki project/fd3f1fcc-1dda-4076-ad4e-7b885ec90032/p... ...
# ovo_qki_0045_cycle04 qki project/fd3f1fcc-1dda-4076-ad4e-7b885ec90032/p... ...
# ovo_qki_0126_cycle01 qki project/fd3f1fcc-1dda-4076-ad4e-7b885ec90032/p... ...
# ovo_qki_0193_cycle01 qki project/fd3f1fcc-1dda-4076-ad4e-7b885ec90032/p... ...
# Get wide descriptor table for the designs
values = descriptor_logic.get_wide_descriptor_table(design_ids=designs.index)
values = designs.join(values) # join with metadata from designs table
values.head()
# pool_id ... Radius of gyration Rosetta ddG
# id ...
# ovo_qki_0045_cycle01 qki ... 75.842162 -47.569473
# ovo_qki_0045_cycle02 qki ... 75.842162 -46.606552
# ovo_qki_0045_cycle04 qki ... 75.842162 -48.614517
# ovo_qki_0126_cycle01 qki ... 65.733242 -33.502827
# ovo_qki_0193_cycle01 qki ... 51.979126 -20.721975
# Show available descriptors
print(values.columns.tolist())
# ['pool_id', 'structure_path', 'structure_descriptor_key', 'accepted', 'spec', 'contig_index', 'Sequence A', 'AF2 iPAE', ...]
# Paths to PDB files are also stored as descriptors:
paths = values[[
"RFdiffusion backbone design",
"ProteinMPNN FastRelax sequence design",
"AlphaFold2 Initial Guess prediction"
]]
paths.head()
# RFdiffusion backbone design ...
# id
# ovo_qki_01_01_cycle01 project/b8e657bb-b0b0-423e-9235-383a6d8f74e5/p ...
# ovo_qki_01_11_cycle02 project/b8e657bb-b0b0-423e-9235-383a6d8f74e5/p ...
# ovo_qki_01_14_cycle01 project/b8e657bb-b0b0-423e-9235-383a6d8f74e5/p ...
# ovo_qki_01_15_cycle02 project/b8e657bb-b0b0-423e-9235-383a6d8f74e5/p ...
# ovo_qki_01_21_cycle03 project/b8e657bb-b0b0-423e-9235-383a6d8f74e5/p ...
# ovo_qki_01_23_cycle02 project/b8e657bb-b0b0-423e-9235-383a6d8f74e5/p ...
See Jupyter notebooks examples for more detailed examples of interacting with the database.